Data from a multiplex output can be used as the quantitative mean fluorescence intensity (MFI) value or as a binary seropositive or seronegative value. A seropositive result can be interpreted as positive for exposure to or infection with a pathogen, or a certain level of seroprotection generated by vaccination depending on the antigen.
Multiplexed serosurveillance assays can efficiently provide information about serological responses to multiple antigens, but these assays also have their challenges for data analysis.
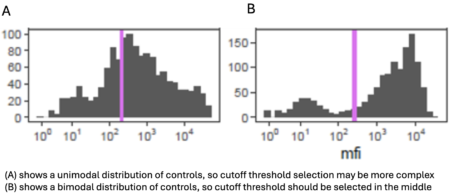
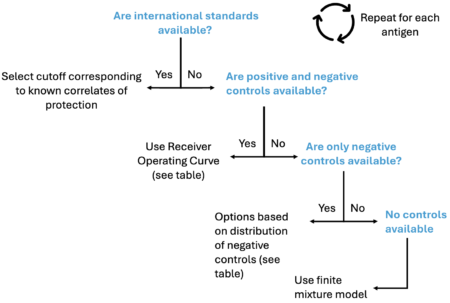
 Positive and negative controls must be identified for each antigen (and those controls must be comparable to the study population), and study plates must include controls for all antigens to determine whether interplate variability could affect study results.
Positive and negative controls must be identified for each antigen (and those controls must be comparable to the study population), and study plates must include controls for all antigens to determine whether interplate variability could affect study results.
 Results for each antigen must be considered individually, and analyses can account for antigen kinetics and whether seropositivity could be affected by vaccination or cross-reactive responses.
Results for each antigen must be considered individually, and analyses can account for antigen kinetics and whether seropositivity could be affected by vaccination or cross-reactive responses.
![]() More complex analyses can also take advantage of results from multiple antigens together, either from the same or different pathogens, to build a more complete antigenic profile of subjects and of the study area.
More complex analyses can also take advantage of results from multiple antigens together, either from the same or different pathogens, to build a more complete antigenic profile of subjects and of the study area.
 Analyses can also incorporate other variables, such as age, to generate seroprevalence curves by age.
Analyses can also incorporate other variables, such as age, to generate seroprevalence curves by age.